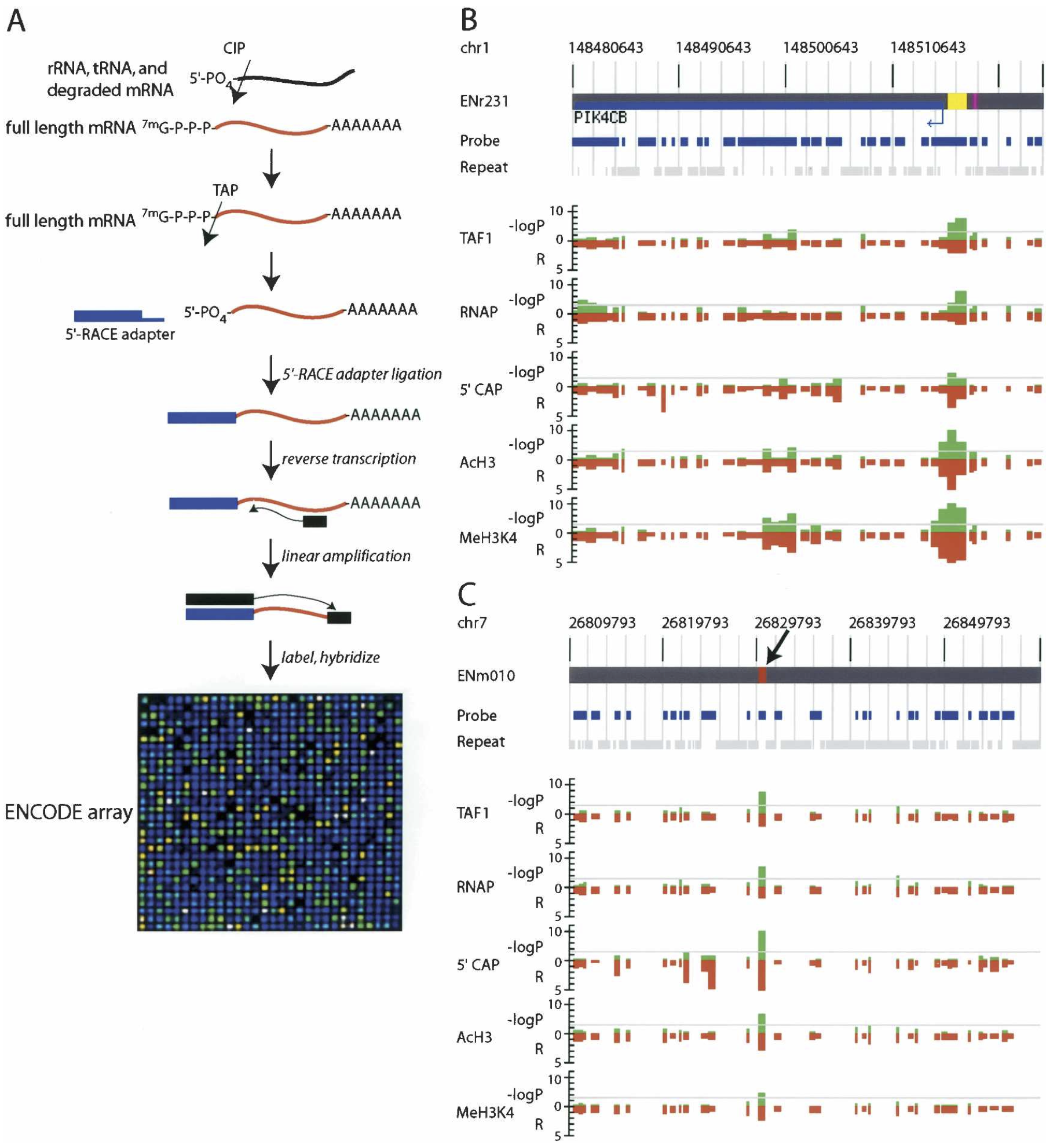
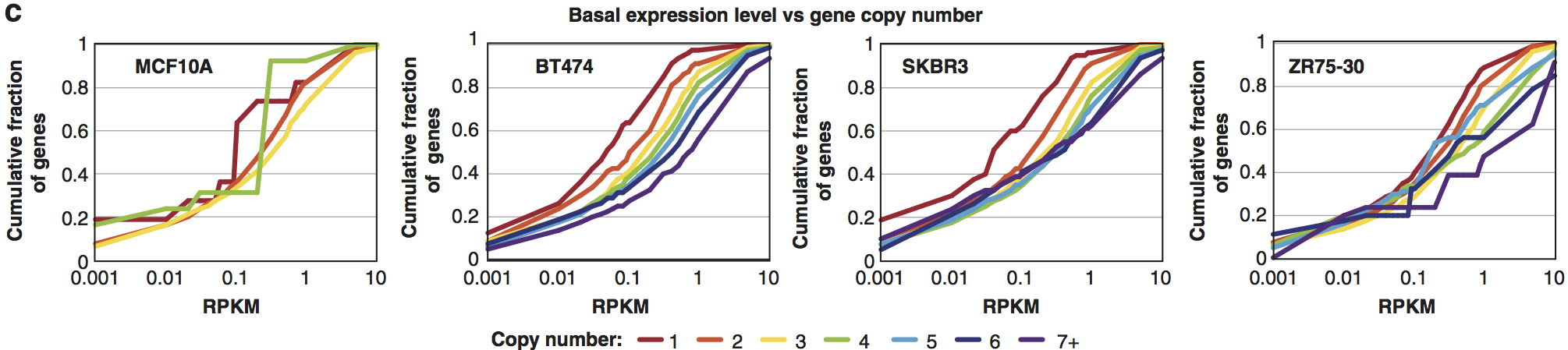
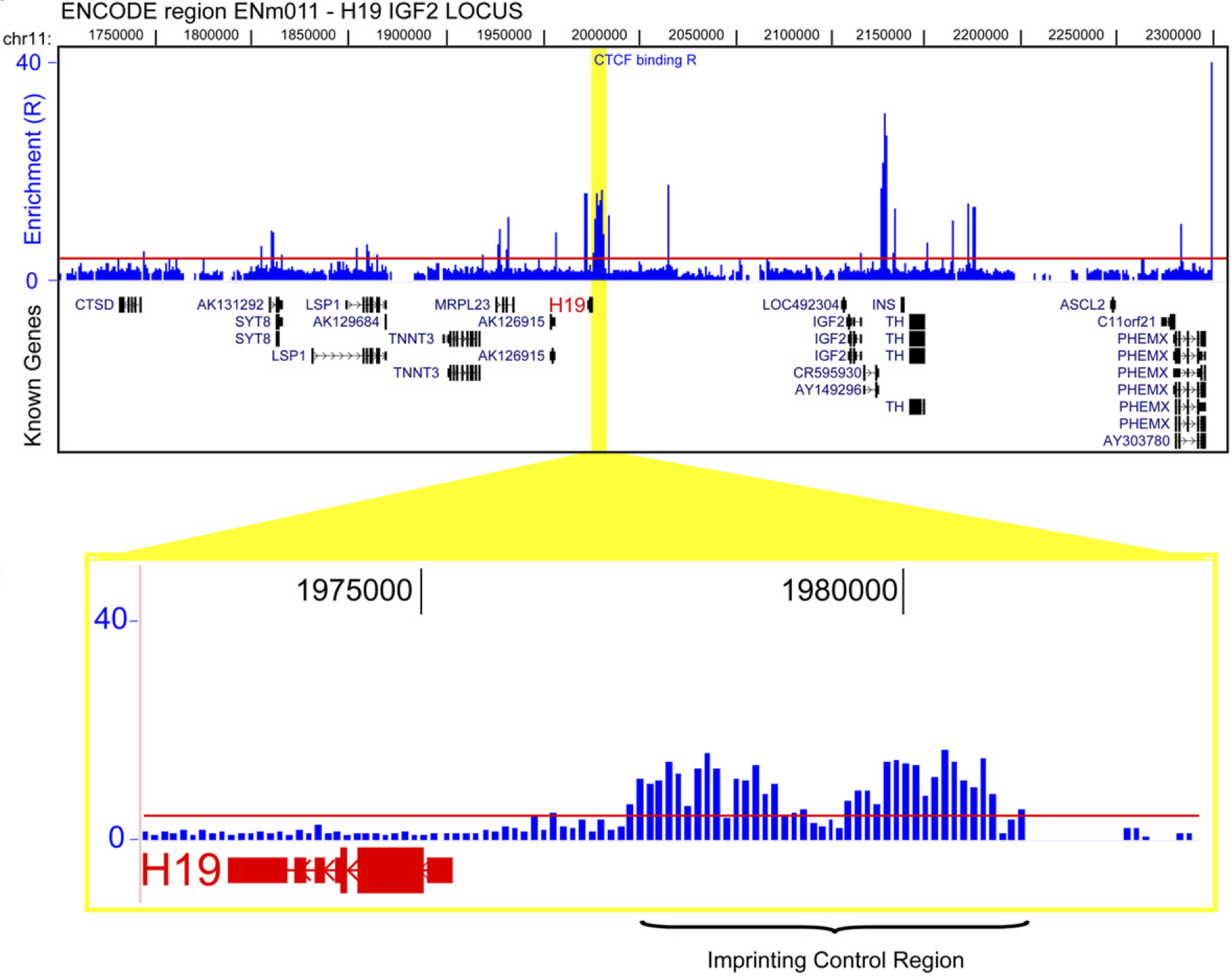
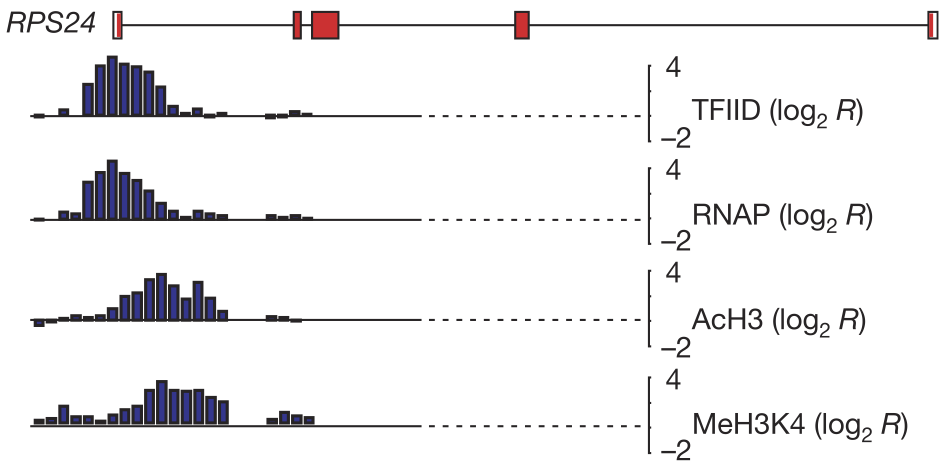
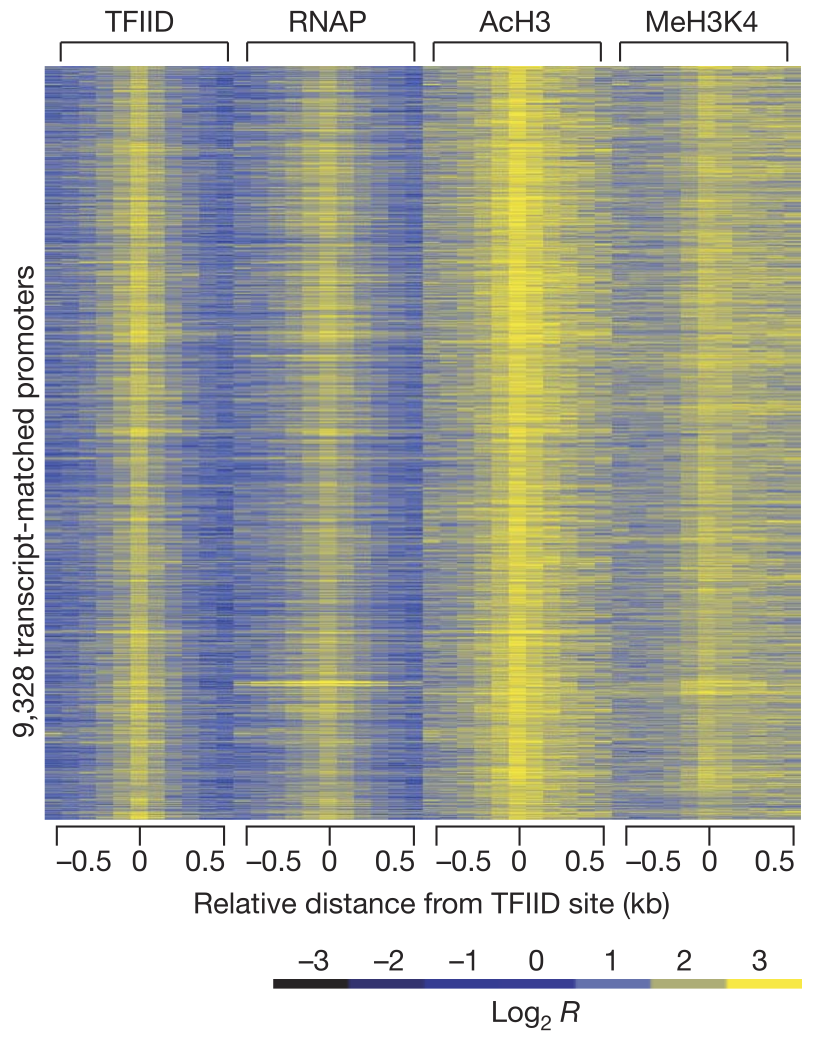
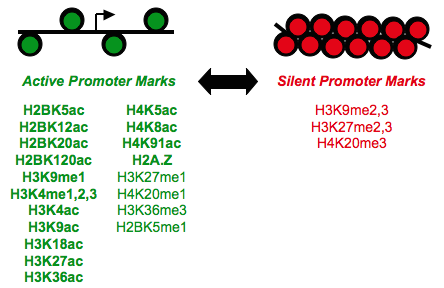
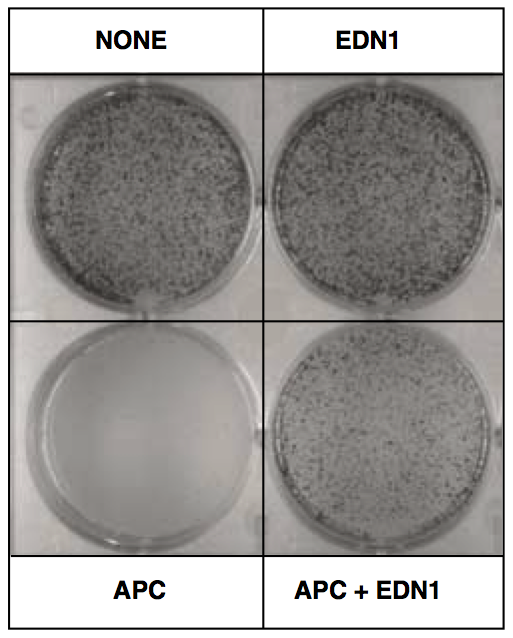
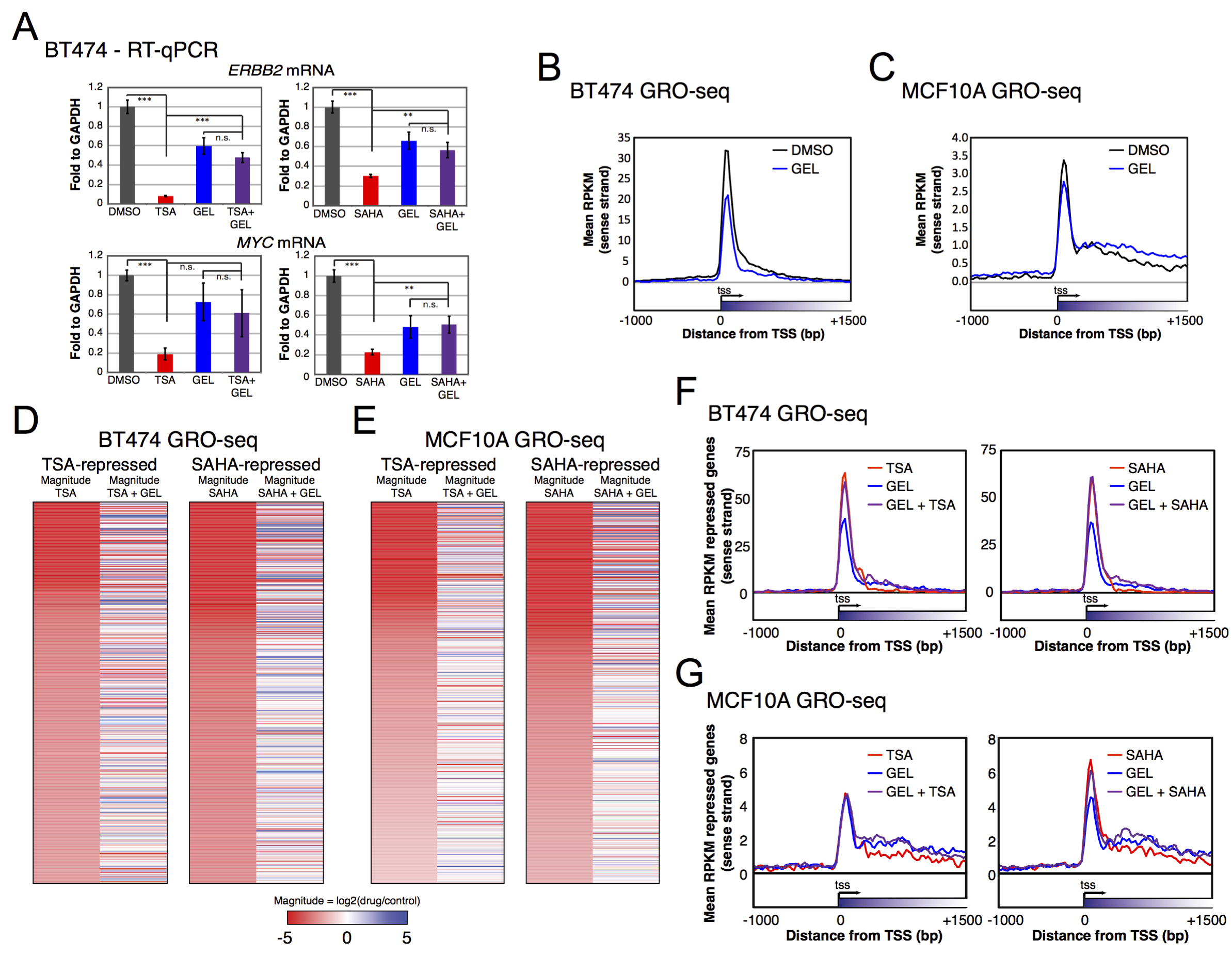
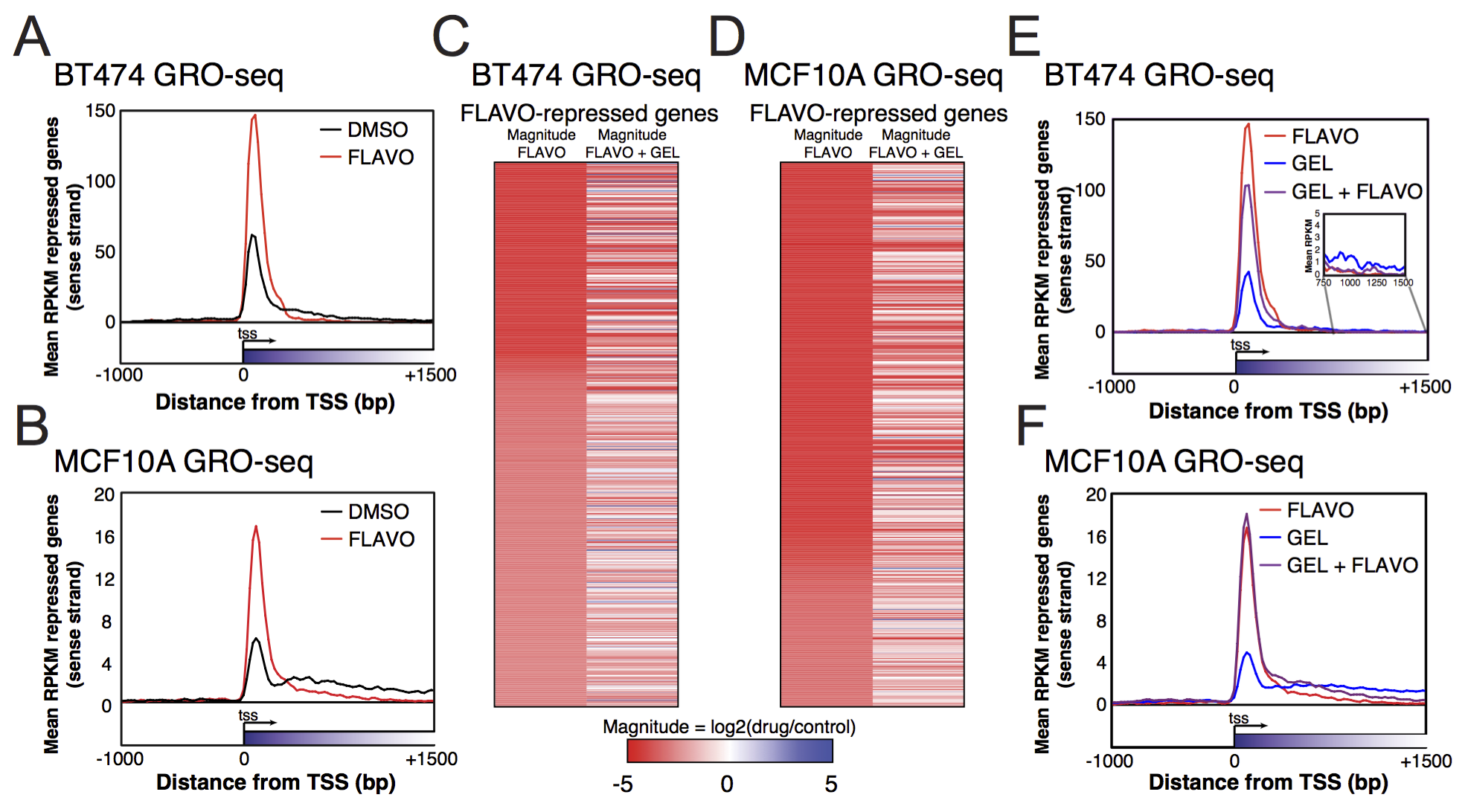
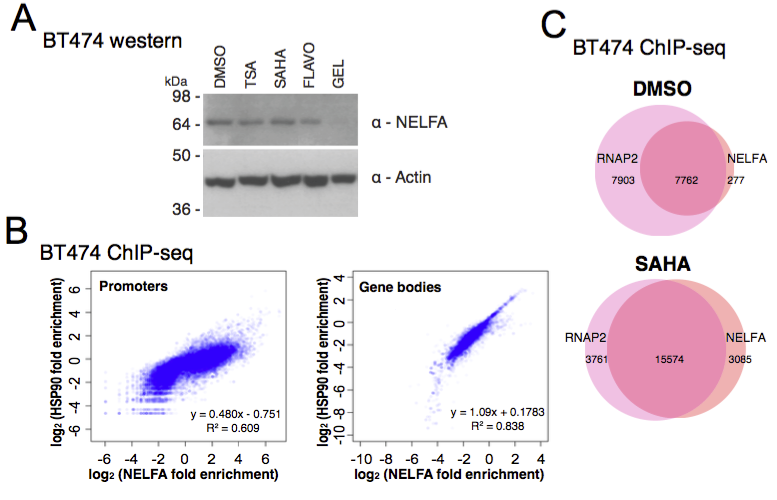
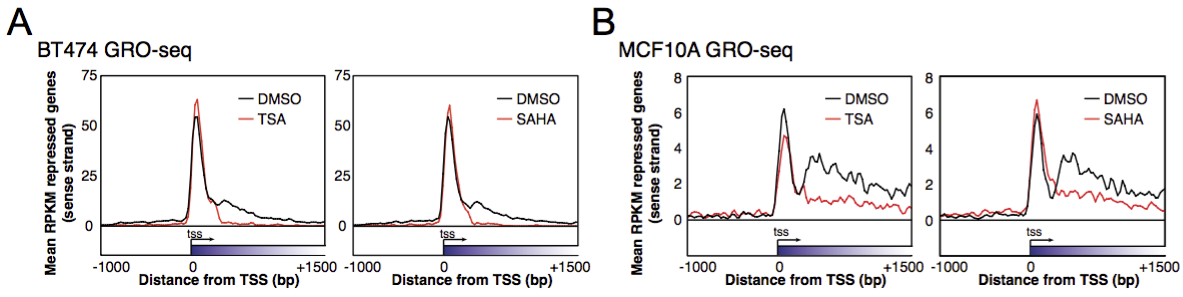
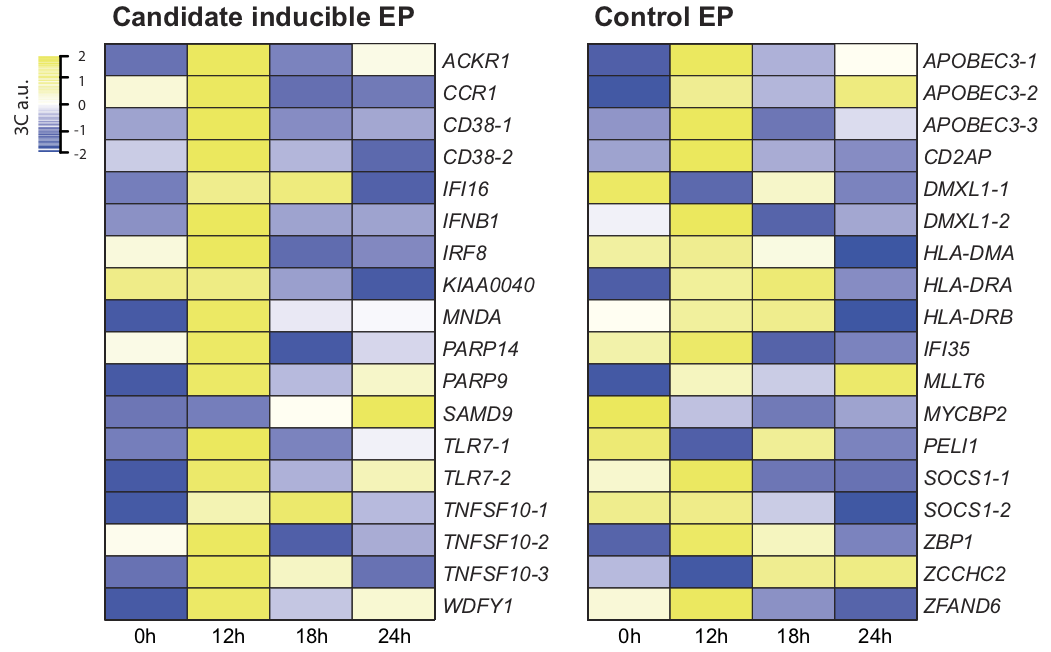
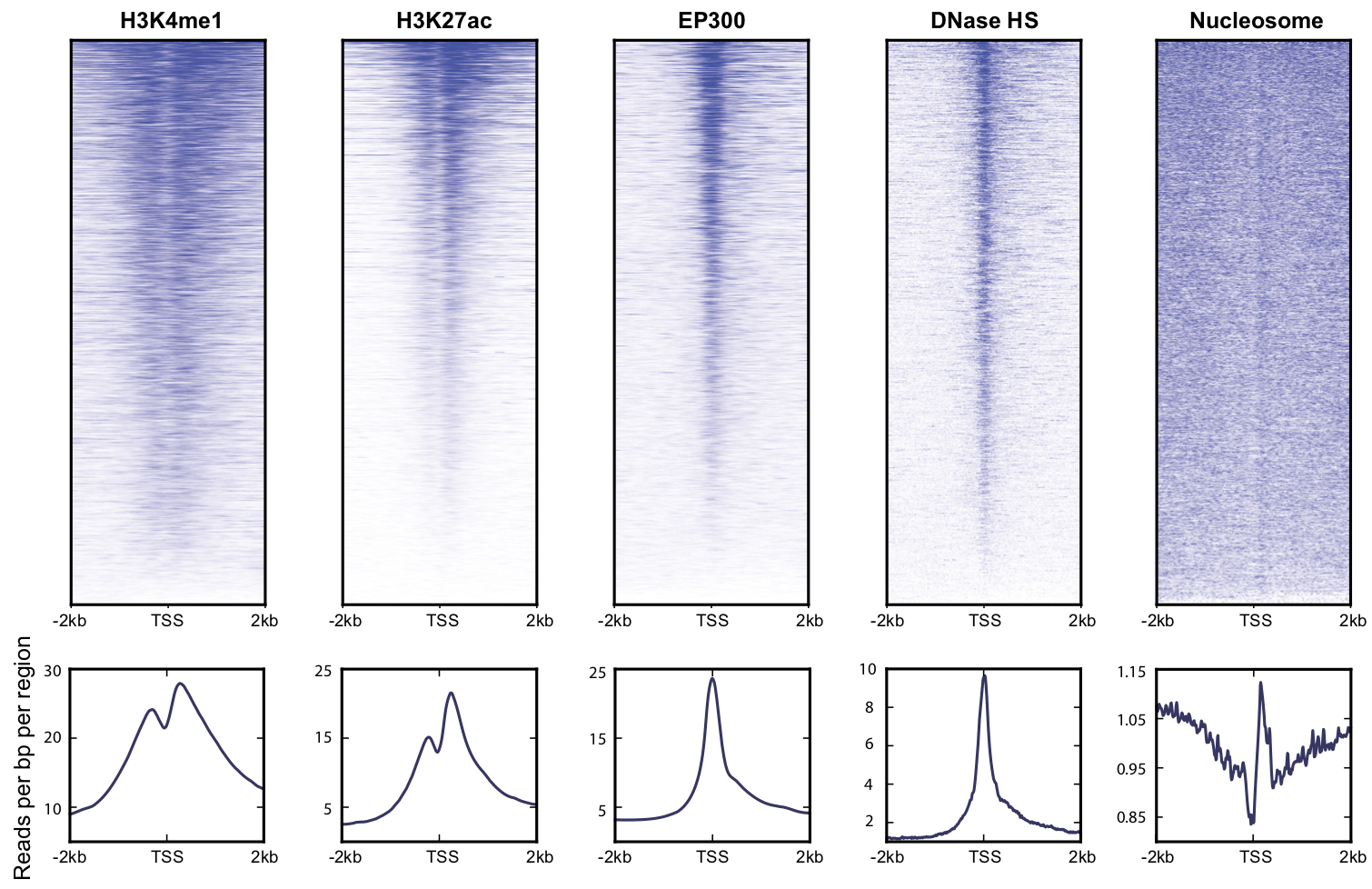
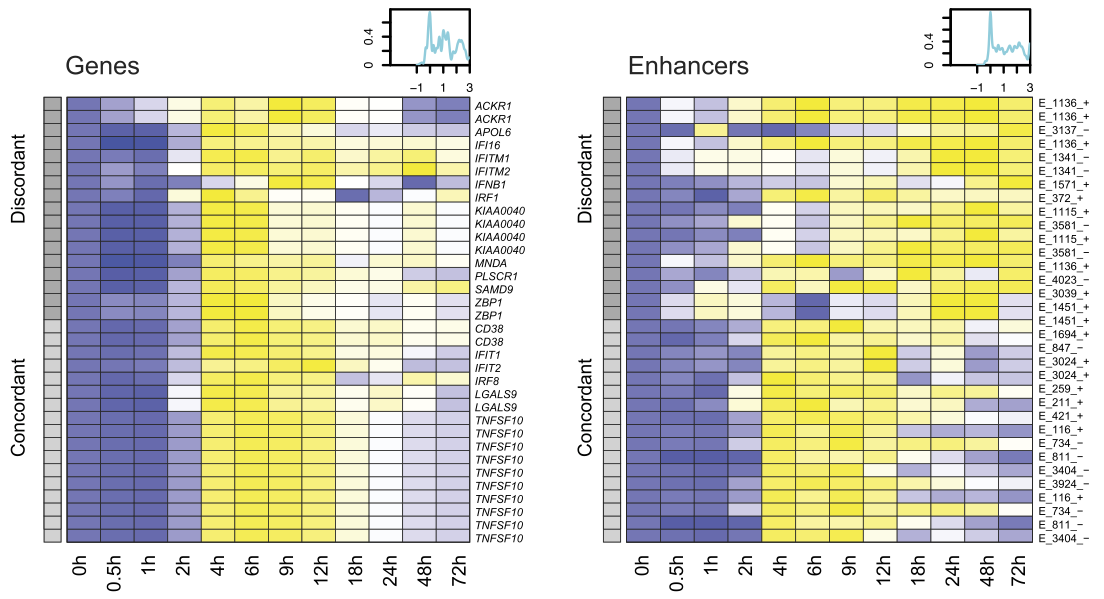
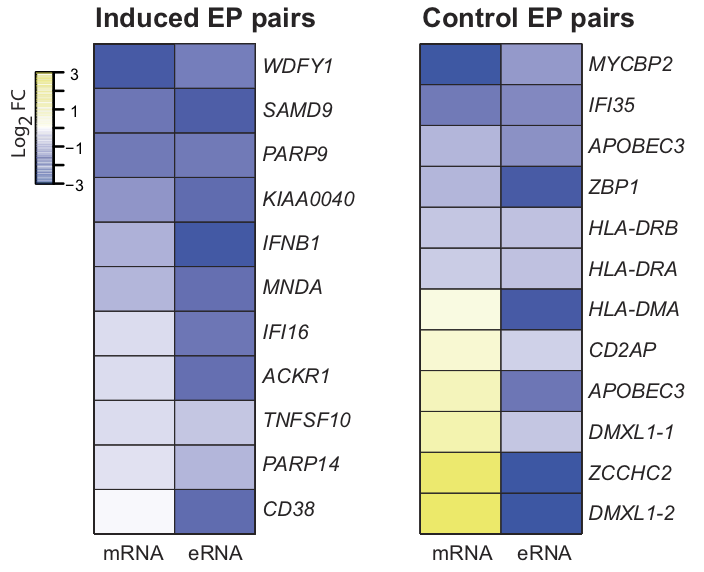
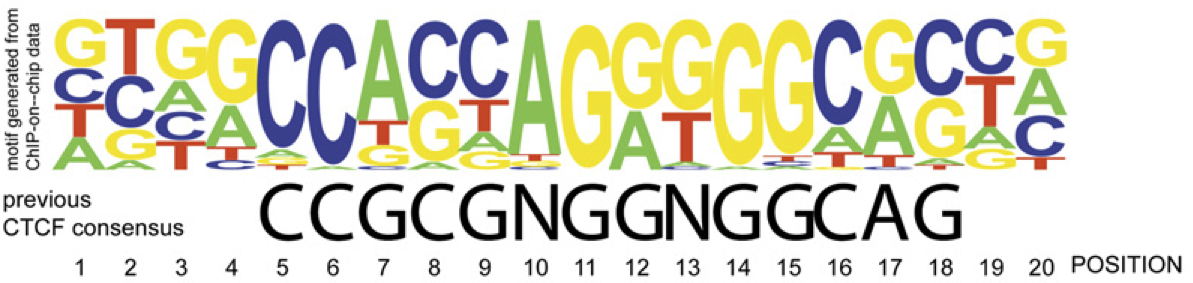View our lablog for major events, milestones and achievements. You can also view our publications and their citations on Google Scholar or PubMed and view/download our genomic data at ArrayExpress. We have a broad interest in genome function and control.
Publications
Tae Hoon Kim. Expression of Salt Tolerance Genes in Vibrio costicola. BASE: A Journal of Science and Technology 8:6-10 (1990).
Tae Hoon Kim. Programmed Cell Death in Arabidopsis thaliana. Reed College. Portland (1994).
Tae Hoon Kim. Structure and Function of Enhanceosome. Harvard University. Cambridge (2002).
Tae Hoon Kim and Bing Ren. All-around view of eukaryotic transcription. Genome Biology 7:323 (2006).
Tae Hoon Kim* and Bing Ren*. Genome-wide Analysis of Protein-DNA Interactions. Annual Review of Genomics and Human Genetics 7:81-102 (2006). *corresponding authors
Tae Hoon Kim*, Leah O. Barrera and Bing Ren*. ChIP-chip for genome-wide analysis of protein binding in mammalian cells. Current Protocols in Molecular Biology 79 Unit 21.13 (2007). *corresponding authors
all genomic data are available at ArrayExpress repository
Tae Hoon Kim* and Job Dekker*. Protein-DNA Interactions. Molecular Neuroscience (ed. Rusty Lansford). Cold Spring Harbor Laboratory Press, Cold Spring Harbor (2014). *corresponding authors
GRO-seq data are available at ArrayExpress repository
GRO-seq and ChIP-seq data are available at ArrayExpress repository
GRO-seq, mRNA-seq and ChIP-seq data are available at ArrayExpress repository
Tae Hoon Kim* and Job Dekker*. Chromatin Immunoprecipitation (ChIP) analysis of protein-DNA interactions. Cold Spring Harbor Protocols doi:10.1101/pdb.top082586 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. ChIP-seq. Cold Spring Harbor Protocols doi:10.1101/pdb.prot082644 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. ChIP-chip. Cold Spring Harbor Protocols doi:10.1101/pdb.prot082636 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. ChIP-Quantitative Polymerase Chain Reaction (ChIP-qPCR). Cold Spring Harbor Protocols doi:10.1101/pdb.prot082628 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. ChIP. Cold Spring Harbor Protocols doi:10.1101/pdb.prot082610 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. Preparation of Cross-Linked Chromatin for ChIP. Cold Spring Harbor Protocols doi:10.1101/pdb.prot082602 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. Formaldehyde Cross-Linking. Cold Spring Harbor Protocols doi:10.1101/pdb.prot082594 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. Generation of 3C Libraries from Cross-linked Cells. Cold Spring Harbor Protocols doi:10.1101/pdb.prot097840 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. Generation of Control Ligation Product Libraries for 3C Analyses. Cold Spring Harbor Protocols doi:10.1101/pdb.prot097865 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. Generation of ChIP-loop libraries. Cold Spring Harbor Protocols doi:10.1101/pdb.prot097857 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. 3C-Based Chromatin Interaction Analyses. Cold Spring Harbor Protocols doi:10.1101/pdb.top097832 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. 4C Analysis of 3C, ChIP-loop, and control libraries. Cold Spring Harbor Protocols doi:10.1101/pdb.prot097881 (2018). *corresponding authors
Tae Hoon Kim* and Job Dekker*. 5C Analysis of 3C, ChIP-loop, and control libraries. Cold Spring Harbor Protocols doi:10.1101/pdb.prot097899 (2018). *corresponding authors
GRO-seq and ChIP-seq data are available at ArrayExpress repository
Boksik Cha, YenChun Ho, Xin Geng, Md Riaj Mahamud, Lijuan Chen, Yeunhee Kim, Dongwon Choi, Tae Hoon Kim, Gwendalyn J Randolph, Xinwei Cao, Hong Chen, and R. Sathish Srinivasan. YAP and TAZ maintain PROX1 expression in the developing lymphatic and lymphovenous valves in response to VEGF-C signaling. Development 147:dev.195453 (2020).
Simbarashe Mazambani, Seong-Ho Park, Joshua Choe, An Nguyen, Bok-Soon Lee, Ji Yun Jeong, Shin Yup Lee, Chul-Ho Kim, Yea-In Park, Joselyn Padilla, Jiyoung Lee, Dinesh Thotala, Tae Gyu Oh, Pankaj Singh, Hoon Hur, Junho Hur, Jung-whan Kim, and Tae Hoon Kim. Targeting the glucose-insulin link in head and neck squamous cell carcinoma induces cytotoxic oxidative stress and inhibits cancer growth. Cancer Research Communications in press (2025).